Page 2 :
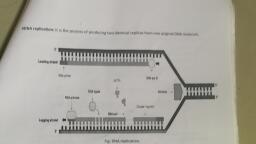
ers believed that gyrase functioned both for helic, , various enzymes both the strands of DN ., “| ‘, , pried ee in | one stretch due to very high energy requirement. The point of, , separation Pp ; — Slowly towards both the directions, In each direction, it gives the, , appearance of Y-shaped structure called replication fork (Fig. 6.13 and 6.14)., , ase and topoisomerase). With the help of, A become open for replication. However, whole of _, , , , » DM, a es, , POLYMERASE e, , , , , , , , , , , , , , , , , , , , , , , , , , , , TOPOISOMERASE, , , , , , , , , , , , , , , , , , , , x, A §sB 5/ PRIMER GAP, REPAIRED, ., 3", ssB ', HELICASE or e, / RELEASED, “Tl il ] \, PRIMER *, 3! \ § a RELEASED, fb DNA POLYMERASE, TOPOISOMERASE Rexg 3!, ', B 5, 1 PARENTAL, OVERALL DIRECTION t 3° STRAND, ', TION, OF REPLICA’ LEADING, STRAND *, , SSB, , , , , , , , ., , , , , , , , , , , , , , , , eo, , t, T &, at \ > & QRAZAA FPAQWENTE_ ul N, 5 , GR LIGASE yu a, , , LAGGING, 3! STRAND, , 5' PARENTAL, ~ STRAND, , inuous over one strand and discontinuous over other strand., , , , — conti, Fig. 6.14. Replication of DNA— con, , , , ; initiati DN ins. RNA primer is a small, : ; tial for initiation of new DNA chains. RNA prin a, , 3. RNA Primer. Itis eed atthe 5’ end of new DNA strand with the help of DNA, strand of RNA which is sy came called rimase. RNA primer is formed on the free end of, , specific RNA polymerase £n2¥" _, , Scanned with CamScanner
Page 3 :
i, , one strand and fork end of the other strand. Formation of RNA primer consti, , z i ce hi . tutes the, initiation phase of DNA synthesis_because_ without the presence of RNA primer, DNA, polymerases cannot add nucleotides. A more complex enzyme called primosome is Tequired |, , in phage @ x 174 and some other prokaryotic systems. In eukaryotes, the function of Primase, , is carried out by enzyme_DNA polymerase o, It builds up ~10 base RNA and 20-30 base, of DNA (Lewin. 2004). After start of nucleotide chain, RNA primer is removed and the a |, , is filled by DNA polymerase | in prokaryotes and DNA polymerase in eukaryotes,, , 4, DNA Polymerases. Prokaryotes have three major types of DN/ synthesising ca, , zymes called DNA polymerases III, II and I. AILof them add nucleotides in 5’-53’ direction, , ~ on 3’ > 5’ stretch of parent strand. They also possess 35! exonuclease ‘activity. While, DNA polymerase III is mainly involved in DNA replication (addition and polymerisation of |, new bases), polymerase I is major repair enzyme. Polymerase II is minor repair enzyme,, DNA polymerase I also has 5 — 3 exonuclease activity. In eukaryotes five types of DNA, polymerases are found— o,, B, y, 5, and ¢, but the major three being «*, 8 and €. Polymerase, § is involved in replication of leading strand. Polymerase e** may help in synthesis of lagging, strand alongwith other roles. Polymerase a is largest and main enzyme of replication of, DNA. All DNA polymerases have a configuration of gripping hand with thumb on one side,, fingers on the other and the palm like concave catalytic site for combining template and base, pairs., , 5. Base Pairing. The two separated DNA strands in the replication fork function as, templateS Deoxyribonucleoside triphosphates come to lie opposite the nitrogen bases of, exposed DNA templates — deTTP opposite A, deCTP opposite G, deATP opposite T and, deGTP opposite C. Nucleophilic attack separates a pyrophosphate (PPi) from the triphosphate. Phosphodiester linkages are established. Hydrolysis of pyrophosphate by enzyme, pyrophosphatase releases energy., , Deoxyribonucleoside triphosphate ———> Deoxyribonucleoside monophosphate + PPi, , PPi + H,O pyrophosphatase, The energy is used in establishing hydrogen bonds between the free nucleotides and, nitrogen bases of templates., , 6. Chain Formation. It requires DNA polymerase III (Kornberg, 1956) in prokaryotes, and polyiiférasé 6 /e in eukaryotes, DNA polymerase IIT is a complex enzyme having seven, subunits (a, B, 8, 7, €, 8, 7). In the presence of Mg?*, ATP (GTP), TPP and DNA polymer, IM], the adjacent nucleotides found attached to nitrogen bases of each template DNA strat, establish phosphodiester bonds and get linked to form replicated DNA strand. As replication, Proceeds, new areas of parent DNA duplex unwind and separate so that replication proces :, rapidly from the place of origin towards the other end. RNA primer is removed and ne, filled with complementary nucleotides by means of DNA polymerase I. Because of ee, wee DNA double chain and its replication to form two chains, DNA replication a in, 55 oe san. However, DNA-polymerase can polymerise nucleotides on ;, , ction on 3’ ~ 5’ strand because it adds them at the 3’ end. Since the two $, “a= alpha § = delta e=, , ‘9 \ : ilon, Previously it was held ei, DNA synthesis) tara th PNA Polymerase 8 (delta) synthesizes the DNA on the leading Sr, , DNA synthesis). Renae DNA polymerase a (alpha) synthesizes DNA on the lagging strand te leading am, , — Pi + Pi + energy, , d (continuous, , Scanned with CamScanner
Page 4 :
119 U2, , MOLECULAR BASIS OF INHERITANCE, , of DNA run in antiparallel direct: icati, , Replication we a Be directions, the {Wo templates provide different ends for replication., ‘ ity J 5:5! Gs vo templates thus Proceeds in opposite directions. One strand with, poli J 38 PMS its © . . : 5 or is, Ee at a complementary strand continuously because 3’end of the latter ts, , always open for elongation Tt is ce ?, : ith pe “eS called leading strg eplication is discontinuous on the, other template with polarity 5” ok: a ng strand. Replication is disc, , ay se? ee de : because only a short seement of DNA strand can be, built in 5 > 3 direction due to exposure of a nea ca ceeie at one time. Short, segments a replicated , DNA are called Okazaki fragments = Okasaki segments ; Reiji, Okazaki. J 968). Each ot them has 1000- 2000 bp in prokaryotes and 100-200 bp in eukaryotes An BNA primer is also required every time a new Okazaki faanient is to be built. After, replacing RNS primer with deoxyribonucleotides and their polymerisation, Okazaki fragments are joined together by means of enzyme, DNA ligase (Khora 1967). DNA strand, built up of Okazaki fragments is called lagging strand.” s, , , , , , , , , , , , As one Strand sows continuously while the other strand is formed discontinuously,, I replication is semidiscontinuous, Since replication proceeds bidirectionally from the, origin of replication or ori, one parent strand will form a leading strand on one side and, laggi strand on the other side. The reverse occurs on the parent strands of the other side., helps in completing replication simultaneously in the whole replicon., , , , , , , , Differences Between Leading Strand and La, , , , , , , Leading Strand eging Strand, , , , , , , , , , , 1, It isa replicated strand of DNA which grows 1. Lagging strand is a replicated strand of, , { continuously without an : ' DNA which is formed in short segments, , ! j called Okazaki fragments. Its growth is, discontinuou, , 2. It does not require DNA ligase for its growth. , 2. DNA-ligase is required for joining Okazaki, , | fragments., 3. The direction of growth of the lagging, , 3. The direction of growth of the leading strand, strand is 3° > 5° though in each Okazaki, , is 5° > 3°, , , , fragment it is 5 4. Only a single RNA primer is required. | 4. Starting of each Okazaki fragment requires a, | new RNA,, 5. Formation of laggi, 6. Its template opens in 5, Zi, , and is slower., ~> 3 direction,, , , , , 5. Formation of leading strand is quite rapid., , | 6. Its template opens in 3° S direction. p h i, | 7. Formation of leading — strand _ begins Formation of lagging strand begins a bit, } immediately at the beginning of replication. | later than that of leading strand., , 7. Proof-reading and DNA Repair. A wrong base is sometimes: introduced during, replication. The frequency is one in ten thousand. DNA polymerase IIT is able to sense the, same. It goes back, removes the wrong base, allows addition of Proper base and then, proceeds forward. However, even DNA polymerase Mis unable to distinguish uracil trom, thymine so that it is often incorporated in place of thymine. Such a mismatching is corrected, by means of a number of enzymes., , There is a separate repair mechanism for any damage caused to DNA due to mutation,, UV exposure or mismatching that escapes proof-reading mechanism. A nick or break is, Caused by an endonuclease near the region of repait. DNA polymerase 1 (Kornberg, 1969), Temoves the mismatched or wrong nucleotides if present and synthesises a correct replaceMent by using the intact strand as template. The newly formed segment is scaled by DNA, , ligase,, , Scanned with CamScanner
Page 5 :
oer, , , , , , , , , , whe oh, , _ Prokaryotic: DNA. Replication oe is Sed eae ad, , salve a, , It occurs inside the cytoplasm., There is single origin of replication., , DNA polymerase III carries out both initiation, and elongation., , DNA repair and gap filling are done by DNA, polymerase I., , RNA primer is’ removed by. DNA, polymerase I. ;, Okazaki fragments are large, 1000-2000, , nucleotides long., , Replication is very rapid, some 2000 bp per, second., , DNA gyrase is needed., , , , wn, , , , , , , oil Ta ication’, , It occurs inside the nucleus., Origin of replications are numerous. ig, Initiation is carried out by DNA polymerase , a while elongation by DNA polymefnse, by, and €., , The same are, polymerase 3. RNA - primer is, polymerase 3., Okazaki fragments are short,, nucleotides long., , Replication is slow, some 100 nucleotides., per second., , DNA gyrase is not needed., , perfanied ny DNA, removed by DNA’ |, , 100-200 °°, , Scanned with CamScanner